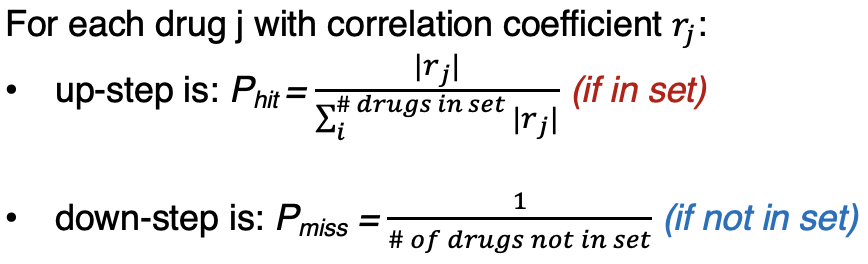Methods
The DMEA algorithm is similar to that of Gene Set Enrichment Analysis (GSEA).
Calculations of Parameters for DMEA:
Enrichment Score (ES): Maximum deviation from 0 on walk down drug list (using weighted, running sum Kolmogorov–Smirnov-like statistic). The score increases if the drug is in the set, or decreases if not. Please refer to the image below to see how the score increments are calculated for drugs in the set (P_hit) or not in the set (P_miss).
Normalized Enrichment Score (NES): ES is divided by the mean of same-signed ES_null based on 1,000 permutations.
p-value: The percentage of same-signed ES_null equal to or greater than the ES divided by the percentage of same-signed ES_null.
q-value: The percentage of same-signed NES_null with NES equal or greater to the observed NES divided by the percentage of same-signed NES equal or greater.
| Parameter | Calculation |
|---|---|
| Enrichment Score (ES) | Maximum deviation from 0 on walk down drug list (using weighted, running sum Kolmogorov–Smirnov-like statistic). The score increases if the drug is in the set, or decreases if not. Please refer to the image below to see how the score increments are calculated for drugs in the set (P_hit) or not in the set (P_miss). |
| Normalized Enrichment Score (NES) | ES is divided by the mean of same-signed ES_null based on 1,000 permutations. |
| p-value | The percentage of same-signed ES_null equal to or greater than the ES divided by the percentage of same-signed ES_null. |
| q-value | The percentage of same-signed NES_null with NES equal or greater to the observed NES divided by the percentage of same-signed NES equal or greater. |