How to Use: Web App
The easiest way to use DMEA is with the web application here: https://belindabgarana.shinyapps.io/dmea. Alternatively, you can access our Docker image of our web app here: https://hub.docker.com/r/belindabgarana/dmea. For more information, please watch our tutorial video below or Contact Us.
To use the web app, you can either run one of our provided examples or select one of 4 input types:
- CMap L1000 query output .gct file (i.e., “query_result.gct” or “ncs.gct”)
- CMap PRISM query output .gct file (i.e., “ncs.gct”)
- Drug rank list .csv file (drug names in column 1, drug rank metrics in column 2, optional: drug mechanisms of action in column 3; headers included)
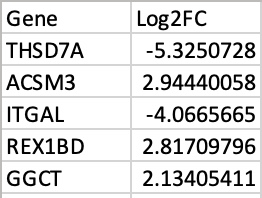
- Gene signature .csv file (gene names in column 1, gene rank metrics in column 2; headers included)
DMEA is flexible to accept any numeric, nonzero values as rank metrics for drugs or genes. To avoid errors, please refresh your web browser after each analysis and format your inputs to match our examples below or on our GitHub repository here: https://github.com/BelindaBGarana/DMEA/tree/shiny-app/Examples. Please note that you will also need to refresh your connection to the web app after 5 minutes of inactivity.
Drug Rank List

If the third column with moa annotations is not provided, our default moa annotations based on the PRISM drug screen dataset will be used (available on our GitHub repository here: MOA_gmt_file_n6_no_special_chars.gmt). Whether moa annotations are provided or not, the moa sets are output with each analysis in a “DMEA_gmt.Rbin” file for any future analyses with our R package for DMEA (described below).
Gene Signature
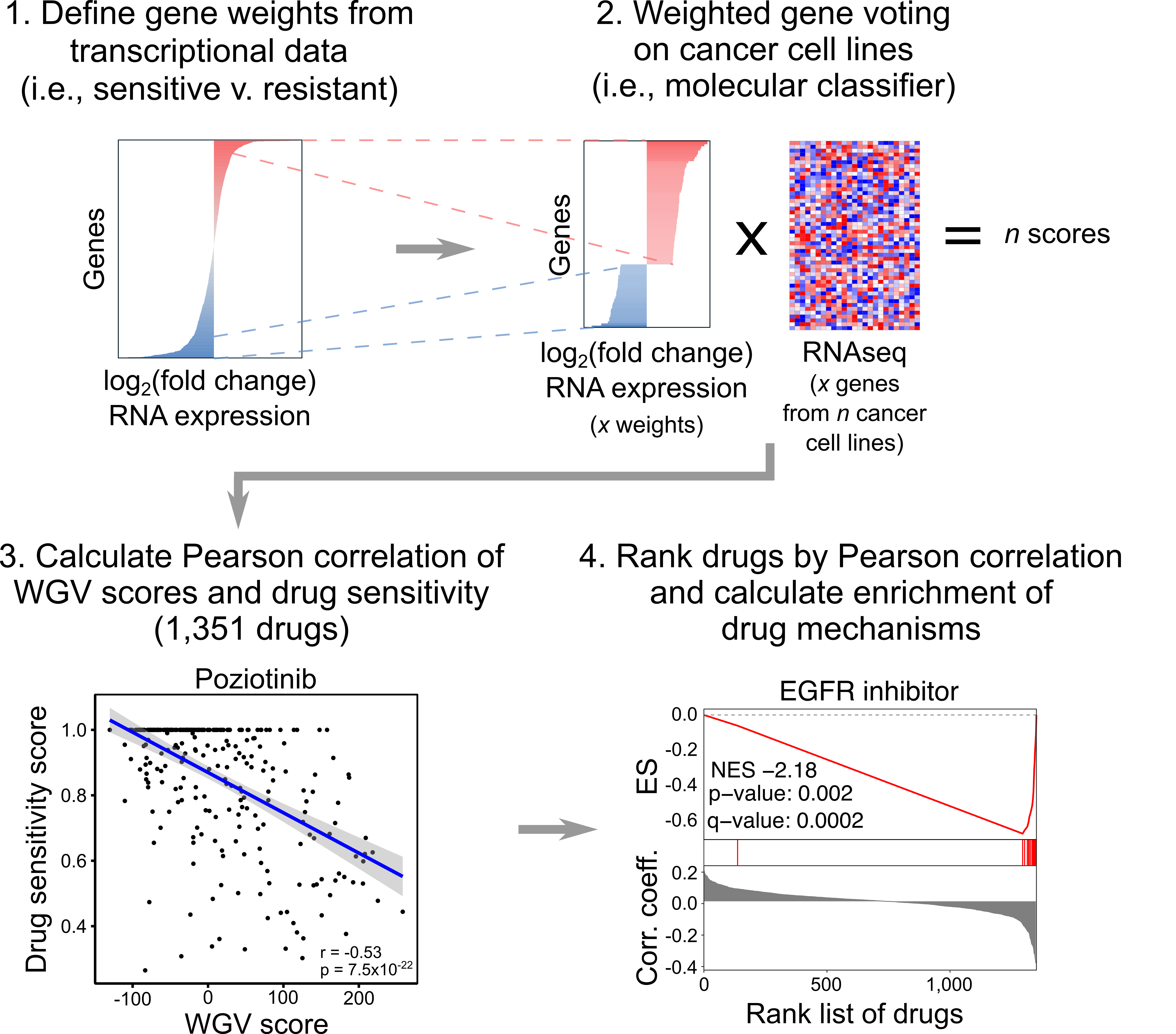
Gene signature inputs will be processed using the PRISM drug sensitivity score metric AUC (Area Under the Curve) and CCLE RNAseq data from version 19Q4.
How to Use: R Package
To use the DMEA R package, first install it by running the 2 lines of code below and then use the steps below to either input a drug rank list or a gene signature. If you use Windows OS, you may need to install the R package using devtools::install_github ('BelindaBGarana/DMEA', build=FALSE). DMEA is flexible to accept any numeric, nonzero values as rank metrics for drugs or genes. To see examples, please refer to the “Examples” folder on our GitHub repository here: https://github.com/BelindaBGarana/DMEA/tree/shiny-app/Examples. More information about how to use our R package is available in our README, vignette, and man files on our GitHub repository: https://github.com/BelindaBGarana/DMEA.
if (!require(devtools)) {install.packages("devtools")}
devtools::install_github ('BelindaBGarana/DMEA')
Drug Rank List
- Install the DMEA R package and load it from your library
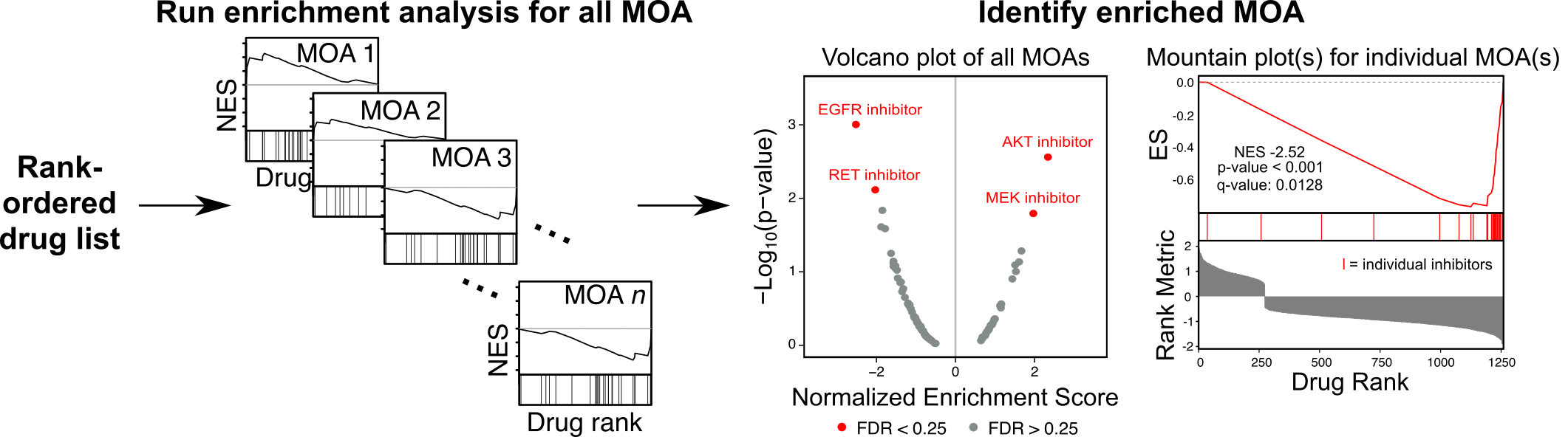
- Input a dataframe containing your drug rank list. If you have mechanism of action (MOA) annotations, you can provide these in a column in your dataframe. If not, you must also input a gmt object such as MOA_gmt_file_n6_no_special_chars.gmt. 2+ MOA must be represented in your input. Make sure each drug has only one numeric, nonzero rank value.
- Save the output dataframe of the enrichment results for each drug set in addition to plots summarizing the results
Gene Signature
- Install the DMEA R package and load it from your library
- Input a dataframe containing your gene signature, as well as dataframes containing drug sensitivity scores and gene expression data with common sample names in the first columns. If you have mechanism of action (MOA) annotations, you can provide these in a separate dataframe. If not, you must also input a gmt object such as MOA_gmt_file_n6_no_special_chars.gmt. This file, in addition to files containing drug sensitivity scores and gene expression data, is available in the "Inputs" folder on our GitHub repository. 2+ MOA must be represented in your input.
- Save the output dataframes with the Weighted Voting, correlation, and enrichment results for each drug set in addition to plots summarizing the results